


Branched TCA Metabolism
Branched Tricarboxylic Acid Metabolism in Plasmodium falciparum.

Diacylglycerol and PIP3 Signaling
Creating cover art and illustrations for MCP, by Rajendrani Mukhopadhyay: ASBMB Today, January 2014
The way Fairman worked on the art for the MCP special issue on posttranslation modi cations was typical for any project she does. She met with Gerald Hart of Johns Hopkins University, the MCP associate editor overseeing the issue, and ASBMB’s publications director, Nancy Rodnan, whose idea it was to hire a professional medical illustrator. Hart explained the science in the various articles. With input from Mary Chang, MCP’s managing editor, the group focused on the images that were either schematics or illustrations. They left alone the images that were captured by a camera or a computer.
“One of the things that I strived to do for this journal was to come up with a consistent style,” explains Fairman. For elements that came up repeatedly, such as ubiquitination, acetylation, proteins and organelles, Fairman established a style so that all of the figures throughout the special issue had the same look and feel. Fairman also says she stuck to scientific conventions as much as possible in terms of colors and symbols. “For example, thinking back to my time in organic chemistry in undergrad, in the little molecular model set, oxygen is usually red, carbon is black, and hydrogen is white,” she says. “Whenever we create any visual, we have to keep in mind who the audience is. Because MCP has a scientifi c audience, I’ve tried to come up with conventions that people are used to seeing.”
Fairman says it can be a challenge to figure out what should be kept in and left out of an illustration. She had a difficult case with one of the figures from the MCP special issue. “ e illustration shows a really complicated mechanism, where these different proteins on the cell membrane, endoplasmic reticulum, nucleus, all the different organelles, are interacting with each other,” she says. “Instead of showing every single protein in its correct configuration, the best thing to do to drive home the message is to use color coding. Not worry so much about what those proteins actually look like but focus more on what they do.”
With the cover, Fairman took another tack, because the cover has a different role than figures in the scienfiti c articles. The inspiration for the cover art came from figure 1 in the article by Corina Antal and Alexandra C. Newton at the University of California, San Diego, on the dynamics of lipid second messenger phosphorylation. “ e cover isn’t necessarily meant to show the whole mechanism in a way that the readers will completely understand it,” says Fairman. “It is supposed to engage them and bring them into the journal, wanting to read that featured article.”
Acknowledgements: Sourceforge Qutemol

Ftz-Z Ring
Ftz-Z Ring
Awarded Ralph Sweet Member’s Choice Award, Traditional Media at the AMI 2015 Annual Conference
Z-Ring Stabilization and Constriction Rate Modulation of the ZapA-ZapB-MatP Protein Network
Artist: Jennifer E. Fairman, CMI, FAMI
Client: Jie Xiao, PHD, Associate Professor Department of Biophysics and Biophysical Chemistry, JHUSOM
Medium / software used: Adobe Illustrator CC, Adobe Photoshop CC and PDB
Final presentation format: 2-Page inside spread for Johns Hopkins Magazine
Primary audience: Alumni, Educated Lay Public
About this project: This illustration was created to illuminate the unique structure known as the “Z-ring” and its associated proteins of the E. coli bacterium, which support and regulate cell division. Published as a 2-page spread (in “Artifact”, a feature that highlights striking, attention-grabbing images of new discoveries found or developed at Hopkins), it illustrates the peptidoglycan (PG) protein structure of the divisome and elongasome protein complexes (ZapA-ZapB-MatP). The divisome and elongasome are responsible for PG synthesis during cell division and elongation, respectively. FtsZ assembles into the Z-ring that determines the division plane. FtsA assists in the formation of the Z-ring, recruits downstream division proteins to the Z-ring to generate the divisome that divides the cell, and is involved in coordinating cell wall synthesis during cytokinesis.
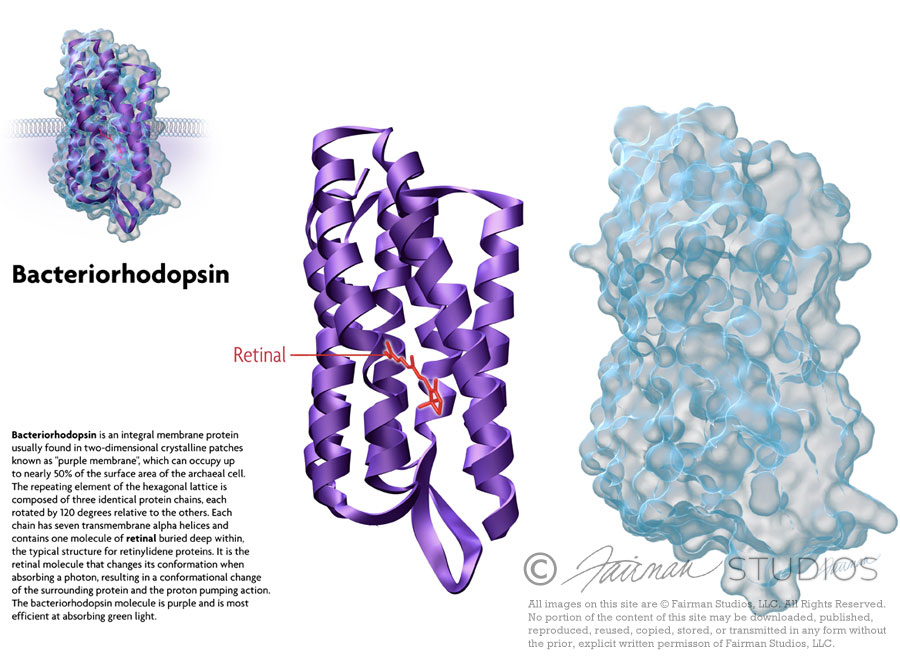
Bacteriorhodopsin
Bacteriorhodopsin is a protein used by Archaea, the most notable one being Halobacteria. It acts as a proton pump; that is, it captures light energy and uses it to move protons across the membrane out of the cell.[1] The resulting proton gradient is subsequently converted into chemical energy.[2]
Bacteriorhodopsin is an integral membrane protein usually found in two-dimensional crystalline patches known as “purple membrane”, which can occupy up to nearly 50% of the surface area of the archaeal cell. The repeating element of the hexagonal lattice is composed of three identical protein chains, each rotated by 120 degrees relative to the others. Each chain has seven transmembrane alpha helices and contains one molecule of retinal buried deep within, the typical structure for retinylidene proteins.
It is the retinal molecule that changes its conformation when absorbing a photon, resulting in a conformational change of the surrounding protein and the proton pumping action.[3] It is covalently linked to Lys216 in the chromophore by Schiff base action. After photoisomerization of the retinal molecule, Asp85 becomes a proton acceptor of the donor proton from the retinal molecule. This releases a proton from a “holding site” into the extracellular side (EC) of the membrane. Reprotonation of the retinal molecule by Asp96 restores its original isomerized form. This results in a second proton being released to the EC side. Asp85 releases its proton into the “holding site,” where a new cycle may begin.
The bacteriorhodopsin molecule is purple and is most efficient at absorbing green light (wavelength 500-650 nm, with the absorption maximum at 568 nm).
Bacteriorhodopsin belongs to a family of bacterial proteins related to vertebrate rhodopsins, the pigments that sense light in the retina. Rhodopsins also contain retinal; however, the functions of rhodopsin and bacteriorhodopsin are different, and there is only slight homology in their amino acid sequences. Both rhodopsin and bacteriorhodopsin belong to the 7TM receptor family of proteins, but rhodopsin is a G protein-coupled receptor and bacteriorhodopsin is not. In the first use of electron crystallography to obtain an atomic-level protein structure, the structure of bacteriorhodopsin was resolved in 1990. It was then used as a template to build models of G protein-coupled receptors before crystallographic structures were also available for these proteins.
Many molecules have homology to bacteriorhodopsin, including the light-driven chloride pump halorhodopsin (for which the crystal structure is also known), and some directly light-activated channels like channelrhodopsin.
All other photosynthetic systems in bacteria, algae, and plants use chlorophylls or bacteriochlorophylls rather than bacteriorhodopsin. These also produce a proton gradient, but in a quite different and more indirect way involving an electron transfer chain consisting of several other proteins. Furthermore, chlorophylls are aided in capturing light energy by other pigments known as “antennas”; these are not present in bacteriorhodopsin-based systems. Last, chlorophyll-based photosynthesis is coupled to carbon fixation (the incorporation of carbon dioxide into larger organic molecules); this is not true for bacteriorhodopsin-based system. Thus, it is likely that photosynthesis independently evolved at least twice, once in bacteria and once in archaea.
Sources:
- Voet, Judith G.; Voet, Donald (2004). Biochemistry. New York: J. Wiley & Sons. ISBN 0-471-19350-X.
- “Bacteriorhodopsin: Pumping Ions”.
- Hayashi S, Tajkhorshid E, Schulten K (September 2003). “Molecular dynamics simulation of bacteriorhodopsin’s photoisomerization using ab initio forces for the excited chromophore”. Biophysical Journal 85 (3): 1440–9. doi:10.1016/S0006-3495(03)74576-7. PMC 1303320. PMID 12944261.
- PDB Molecule of the Month pdb27_1

