Project Description
By Alexander Gelfand, for the Johns Hopkins School of Public Health Magazine.
© Johns Hopkins University
The powerful genome-editing technology known as CRISPR-Cas made headlines this year—partly because many leading biologists called for a moratorium last March against using it to modify the genomes of human embryos, only to discover in April that Chinese scientists had already done just that.
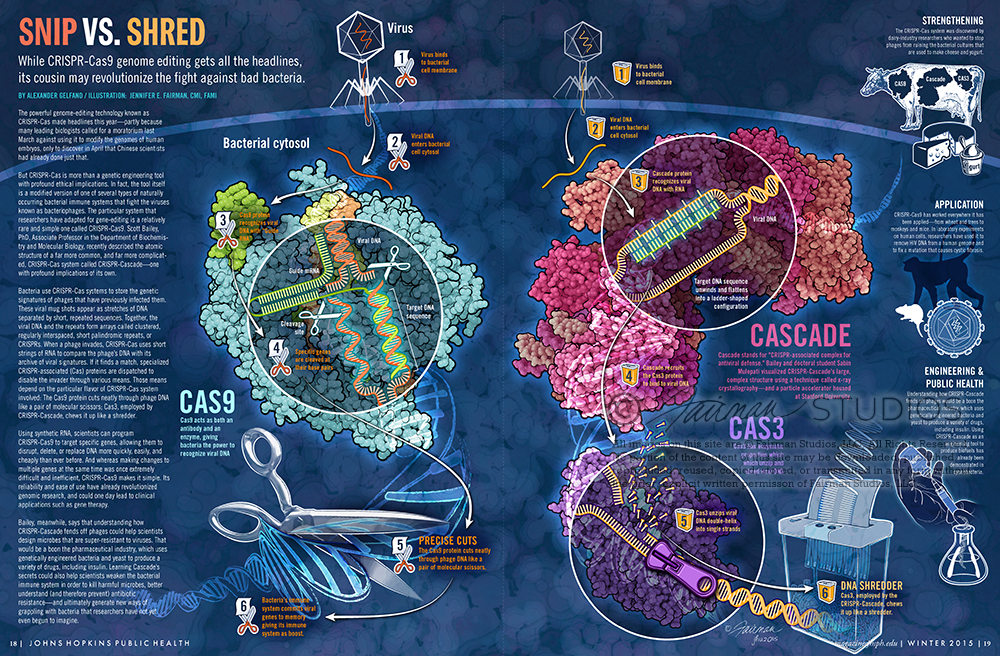
But CRISPR-Cas is more than a genetic engineering tool with profound ethical implications. In fact, the tool itself is a modified version of one of several types of naturally occurring bacterial immune systems that fight the viruses known as bacteriophages. The particular system that researchers have adapted for gene-editing is a relatively rare and simple one called CRISPR-Cas9. Scott Bailey, PhD, Associate Professor in the Department of Biochemistry and Molecular Biology, recently described the atomic structure of a far more common, and far more complicated, CRISPR-Cas system called CRISPR-Cascade—one with profound implications of its own.
Bacteria use CRISPR-Cas systems to store the genetic signatures of phages that have previously infected them. These viral mugshots appear as stretches of DNA separated by short, repeated sequences. Together, the viral DNA and the repeats form arrays called clustered, regularly interspaced, short palindromic repeats, or CRISPRs. When a phage invades, CRISPR-Cas uses short strings of RNA to compare the phage’s DNA with its archive of viral signatures. If it finds a match, specialized CRISPR-associated (Cas) proteins are dispatched to disable the invader through various means. Those means depend on the particular flavor of CRISPR-Cas system involved: The Cas9 protein cuts neatly through phage DNA like a pair of molecular scissors; Cas3, employed by CRISPR-Cascade, chews it up like a shredder.
Using synthetic RNA, scientists can program CRISPR-Cas9 to target specific genes, allowing them to disrupt, delete, or replace DNA more quickly, easily, and cheaply than ever before. And whereas making changes to multiple genes at the same time was once extremely difficult and inefficient, CRISPR-Cas9 makes it simple. Its reliability and ease of use have already revolutionized genomic research, and could one day lead to clinical applications such as gene therapy.
Bailey, meanwhile, says that understanding how CRISPR-Cascade fends off phages could help scientists design microbes that are super-resistant to viruses. That would be a boon the pharmaceutical industry, which uses genetically engineered bacteria and yeast to produce a variety of drugs, including insulin. Learning Cascade’s secrets could also help scientists weaken the bacterial immune system in order to kill harmful microbes, better understand (and therefore prevent) antibiotic resistance—and ultimately generate new ways of grappling with bacteria that researchers have not yet even begun to imagine.
The CRISPR-Cas system was discovered by dairy-industry researchers who wanted to stop phages from ruining the bacterial cultures that are used to make cheese and yogurt.
CRISPR-Cas9 has worked everywhere it has been applied—from wheat and trees to monkeys and mice. In laboratory experiments on human cells, researchers have used it to remove HIV DNA from a human genome and to fix a mutation that causes cystic fibrosis.
Cascade stands for “CRISPR-associated complex for antiviral defense.” Bailey and doctoral student Sabin Mulepati visualized CRISPR-Cascade’s large, complex structure using a technique called x-ray crystallography—and a particle accelerator housed at Stanford University.
Acknowledgements: Sourceforge Qutemol